Target Validation
Target validation is a crucial early step for developing a drug against a new cancer target. There is no universally applicable process for target validation; therefore, our team will create a research plan with the client tailored to the target of interest. When designing the project we account for potential pitfalls such as results confounded by off-target effects.
Target validation at Reaction Biology is mainly based on our suite of cellular and in vivo oncology models. Here are a few examples of target validation approaches:
- Target knockout is one approach to determine if the target is related to the growth or transformation of cancer cells
- Cells with target knockout can be subjected to phenotypic assays such as the cell transformation assay, migration assays, invasion assays, or proliferation assays
- Cells with target knockout can also be investigated in mice to find out if the lack of target expression has any impact on the tumor growth in vivo
Cellular target engagement assays and cellular enzyme activity assays will be performed to determine the drug concentration necessary to reach relevant target occupation or inhibition. This information is crucial for proper interpretation of results with phenotypic assays or tumor models preventing toxicities that originate in too high a dosage.
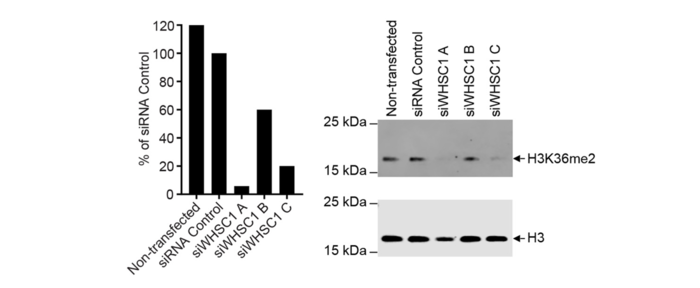
Knockdown of the target can be achieved in three ways:
- Transient knockdown is performed with target-specific RNA interference.
- The well-established Tet-On/Off technology can be employed to modulate target expression.
- Complete knockout is achieved with CRISPR technology. Cell lines with CRISPR-mediated knockdown will be generated by a partner CRO.