MST Assay Service for Drug Discovery
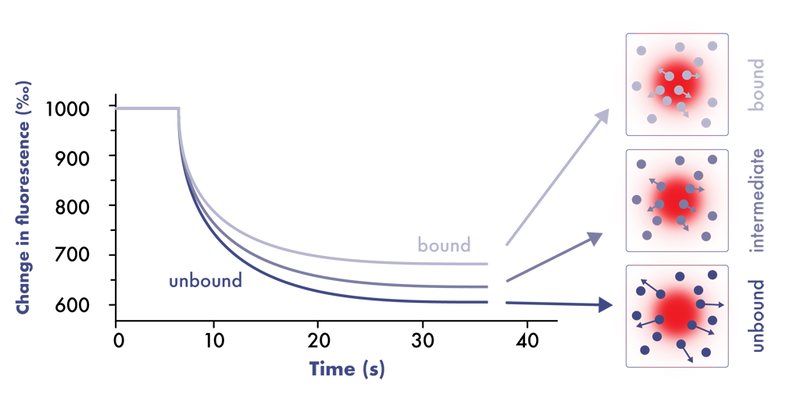
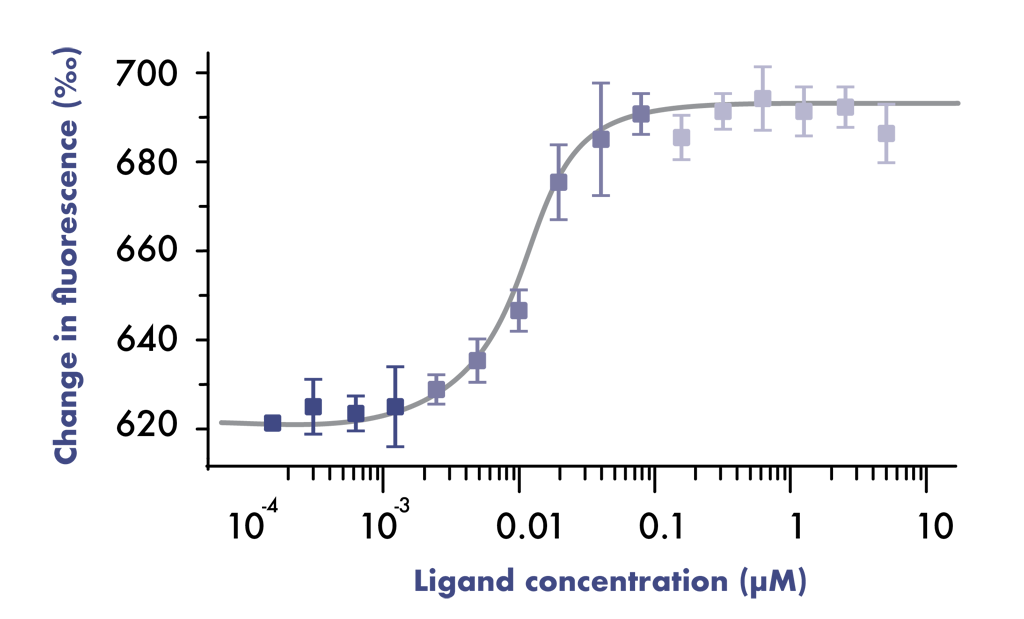
Microscale Thermophoresis assays (MST assays) measure the motion of molecules along microscopic temperature gradients that changes upon ligand binding.
MST assays measure the ligand-target interactions directly in solution without the need for immobilization to a surface (immobilization-free technology). The technique requires low amounts of pure analyte samples and may be compatible with non-purified protein samples and is compatible with most buffers.
Microscale Thermophoresis Assay Methods:
Our MST assay services include:
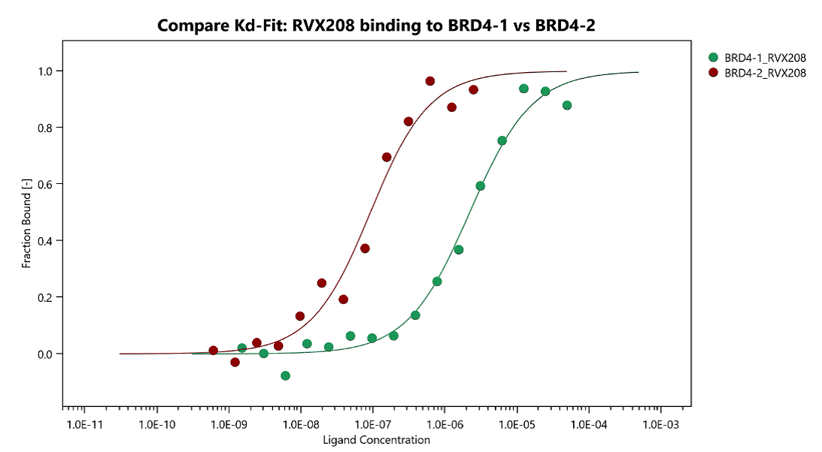
- Binding between biomolecules of any type can be investigated including small-molecules, multi-protein complexes, DNA, liposomes, nano-particles and more
- Fast lead time for assay development and affinity determination
- Deliverable: dissociation constant KD (binding affinity)