Protein-Protein Interaction Assays: Illuminating the Science Behind LinkLight™
By turning fleeting protein-protein interactions into stable, low-noise signals, LinkLight helps discovery teams generate decision-ready data and move programs forward with confidence.
PPI biology balances promise and risk
Protein-protein interactions (PPIs) are at the center of many critical cellular processes. They guide signaling, regulate gene expression, and ensure the smooth coordination of biological pathways. Dysregulation and disruption of these interactions cause numerous diseases with widespread and life-altering implications, from overactive oncogenes in cancer to misfolded proteins in neurodegeneration.
Unsurprisingly, PPIs are attractive targets for novel therapeutics, and understanding them is βbvital for drug discovery and development. For example, roughly 35% of all FDA-approved drugs act on GPCRs,1 where understanding downstream signaling interaction partners, such as β-arrestin and 14-3-3 hub proteins, is key to understanding therapeutic effects.
Moreover, the concept of biased agonism—designing drugs that selectively favor certain signaling pathways—depends entirely on measuring these protein partnerships with specificity. PPIs are also at the heart of emerging therapeutic strategies, such as molecular glues and targeted protein degradation, making their accurate measurement more critical than ever.
Yet PPIs are challenging to capture. Many are fleeting, weak, or occur only inside living cells. Complex signaling pathways may induce multiple PPIs that are difficult to dissect and can be missed by assays with transcription-dependent or transient signal generation. For biotech and biopharma R&D teams tasked with producing reliable, decision-ready data, this makes confident progression a constant challenge.
Inaccurate or incomplete PPI data can send discovery programs down the wrong path, leading to wasted months of development and millions in sunk costs.
That’s why high-quality interaction data, available early in the pipeline, can help drug hunters de-risk decisions and progress programs with confidence. Reaction Biology has over 20 years of experience supporting these needs across our diverse target catalog and assay capabilities, and today, we are adding the LinkLight functional cell-based assay platform for the detection of fleeting PPIs to our discovery portfolio.
From cAMP to β-arrestin: the PPI readouts that matter for GPCR researchers
Researchers studying protein-protein interactions (PPIs) have a range of tools at their disposal—binding assays (radioligand, SPR), proximity-based methods (FRET, TR-FRET), and transcriptional reporters. Yet, each comes with trade-offs in cellular context, geometry/orientation, and signal durability, revealing only part of the full picture. This is especially critical in GPCR drug discovery, where transient and conformation-dependent interactions often determine downstream signaling and therapeutic relevance.
In practice, most GPCR protein-protein interaction assays revolve around three readouts:
- Second messengers (cAMP/Ca²⁺): Fast, high-throughput snapshots of G-protein pathway activation (e.g., Gs/Gi→cAMP; Gq→Ca²⁺). Useful for potency/efficacy and proximal receptor activity on targets with measurable couplings.
- β-arrestin recruitment: A broadly conserved pathway that drives receptor desensitization/internalization and can initiate G-protein-independent signaling. Recruitment and interaction persistence are ligand-dependent, so sensitive live-cell detection is especially helpful for understanding pathway preference.
- 14-3-3 and related adaptor scaffolding: Phosphorylation-dependent interactions that influence receptor trafficking, signal duration, and network crosstalk, valuable when the mechanism and pathway state are the focus.
Comprehensive GPCR functional characterization ideally combines G-protein pathway readouts (cAMP/Ca²⁺) with regulatory mechanism analysis (β-arrestin recruitment), while 14-3-3 interactions provide insights into downstream signal stabilization and crosstalk.
For example, LinkLight can play an important role in your GPCR characterization strategy as a β-arrestin recruitment assay to complement understanding of second messenger cascades. Its protease-triggered design captures split-second arrestin engagement in living cells and converts it into a stable, low-noise signal suited to high-throughput decision-making. Because β-arrestin is engaged across many GPCR classes—including difficult or orphan targets—LinkLight provides a practical, broadly applicable pathway readout for GPCR studies.
How LinkLight fits into the drug discovery toolkit
LinkLight is a functional, cell-based assay that detects fleeting protein–protein interactions in living cells and converts them into a persistent, non-reversible luminescent signal suitable for high-throughput decisions.
The LinkLight mechanism
The LinkLight assay is based on tobacco etch virus (TEV) protease cleavage and luciferase complementation technology. TEV is a small, sequence-specific enzyme that recognizes a defined seven-amino-acid motif and is widely used because of its high specificity and minimal off-target cleavage in mammalian cells.
For the assay, Protein A is fused to TEV protease, and Protein B is fused to a permuted luciferase (pLuc) interrupted by TEV recognition/cleavage sequences. When A and B interact inside the cell, TEV is brought into proximity with pLuc and cleaves at its recognition site. Cleavage allows the luciferase fragments to refold into an active enzyme. In the presence of luciferin, the enzyme emits light. Because TEV cleavage is irreversible, the signal persists even after the proteins separate, providing a “molecular memory” of transient events.
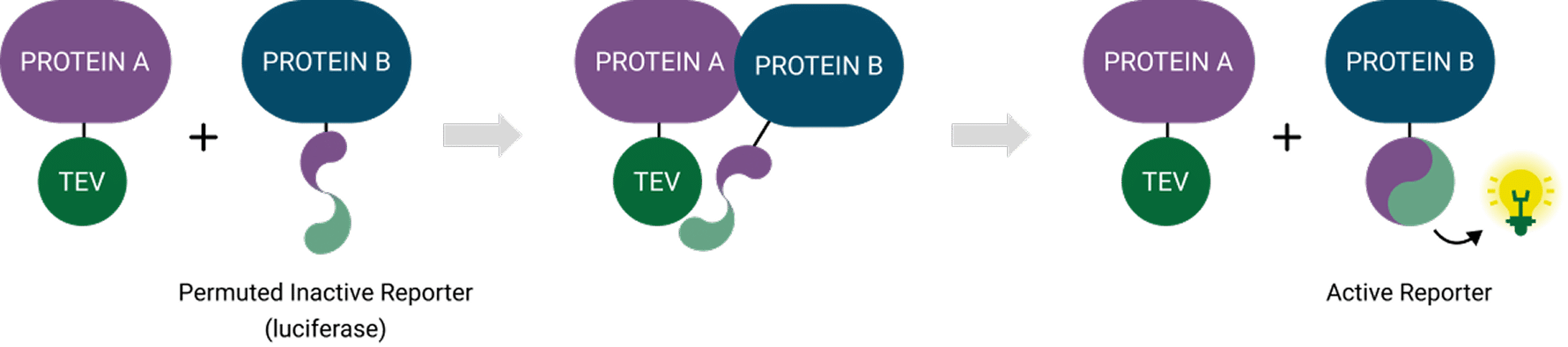
How it maps to GPCR β-arrestin recruitment
When using LinkLight for GPCR assays, the GPCR (Protein A) carries TEV protease, and β-arrestin (Protein B) carries the permuted luciferase. Upon ligand activation, the receptor recruits β-arrestin, bringing TEV next to pLuc, and a stable luminescent signal is produced. The integrated signal reflects the cumulative recruitment events over the read window—ideal for when recruitment is brief, weak, or context-dependent.
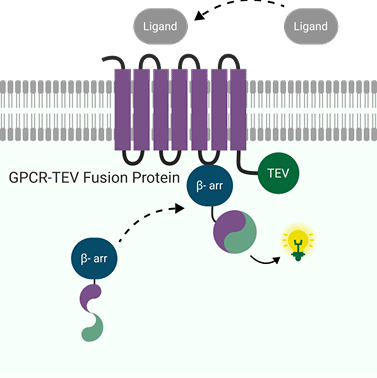
LinkLight provides the unique advantage of generating a durable signal from a fleeting event. Because TEV cleavage is irreversible, even millisecond β-arrestin recruitment leaves a measurable trace (“molecular memory”). The refolded luciferase produces a stable, low-noise light output that holds for hours, making it friendly to automation and flexible read times.
There’s no transcriptional lag—the readout starts at the moment of cleavage—so it’s fast for screening and rapid triage. And because β-arrestin engagement is broadly conserved across activated GPCRs, LinkLight provides a practical, pathway-agnostic readout when you’re working across diverse or hard-to-measure targets, including orphan receptors.
See which cell lines and targets we offer with LinkLight™ PPI assay services
Where LinkLight shines
As part of Reaction Biology’s GPCR discovery portfolio, LinkLight is delivered as a high-throughput, in-lab service with reproducible signals suitable for screening and rapid triage. LinkLight provides sensitive, reproducible signals and fits seamlessly into high-throughput discovery workflows, overcoming the limitations of other protein-protein interaction assays.
Utility as a complementary detection strategy
Using both secondary messenger and β-arrestin assays provides a more complete view of GPCR signaling. Because most GPCRs recruit β-arrestin independently of their G-protein partners, LinkLight β-arrestin assays offer a nearly universal way to monitor receptor activity, while secondary messenger readouts such as cAMP or calcium highlight specific downstream pathways.
Together, these complementary approaches deliver advantages in three key applications:
- Pathway discovery: Detect receptor activity even when GPCR coupling is unknown by combining β-arrestin and secondary messenger assays.
- Ligand bias profiling: Differentiate compounds that favor G-protein vs. β-arrestin signaling, enabling quantification of functional selectivity.
- β-arrestin–specific drug discovery: Develop ligands that minimize G-protein–driven side effects while leveraging β-arrestin’s prolonged and tissue-specific signaling.
This multi-dimensional strategy is particularly valuable for profiling ligand bias, as some compounds preferentially activate G-protein signaling, others β-arrestin signaling, while balanced agonists stimulate both. By quantifying these differences, it becomes possible to detect functional selectivity, guide lead optimization, and better understand mechanisms of action.
Beyond discovery, β-arrestin-biased ligands also hold therapeutic promise by minimizing side effects linked to G-protein signaling, enabling tissue—and cell–type–specific targeting, and supporting longer-lasting therapeutic responses.
What this means for your team’s protein-protein interaction assays
For biotech and biopharma companies, LinkLight directly supports strategic R&D goals:
- De-risk early decisions: Generate reproducible interaction data that reduces the chance of false positives.
- Accelerate milestones: Outsourcing can reduce development timelines from months to weeks, particularly for organizations without existing LinkLight expertise.
- High-quality screening: Stable, low-background signals support confident progression or termination calls.
- Access specialized expertise: Service providers bring validated assay libraries, technical guidance, and quality systems suitable for regulatory submissions.
By integrating LinkLight alongside complementary approaches, discovery programs can progress with greater speed and confidence, while keeping internal capacity focused where it creates the most value.
Turn elusive signals into confident decisions
Protein-protein interaction assays are essential to modern drug discovery but remain among its toughest challenges. The adoption of next-generation approaches like LinkLight reflects the field’s progress in meeting that complexity. LinkLight’s contribution lies in its ability to capture elusive interactions with stable, decision-ready signals. For discovery teams under pressure to de-risk programs, accelerate timelines, and access high-quality data, LinkLight represents a proven, efficient way to move forward with confidence.
At Reaction Biology, our team is always looking ahead. With molecular glues, protein degraders, and other novel therapeutic modalities dependent on understanding and measuring protein partnerships, it’s clear that the importance of PPI assays will only increase. As drug discovery moves deeper into these complex signaling networks, we are excited to provide support with like LinkLight that reliably illuminate these interactions and help determine which programs succeed.
Thinking of including LinkLight in your next GPCR study? Our scientific service team can help strengthen your discovery toolkit and turn elusive protein-protein interactions into confident decisions.