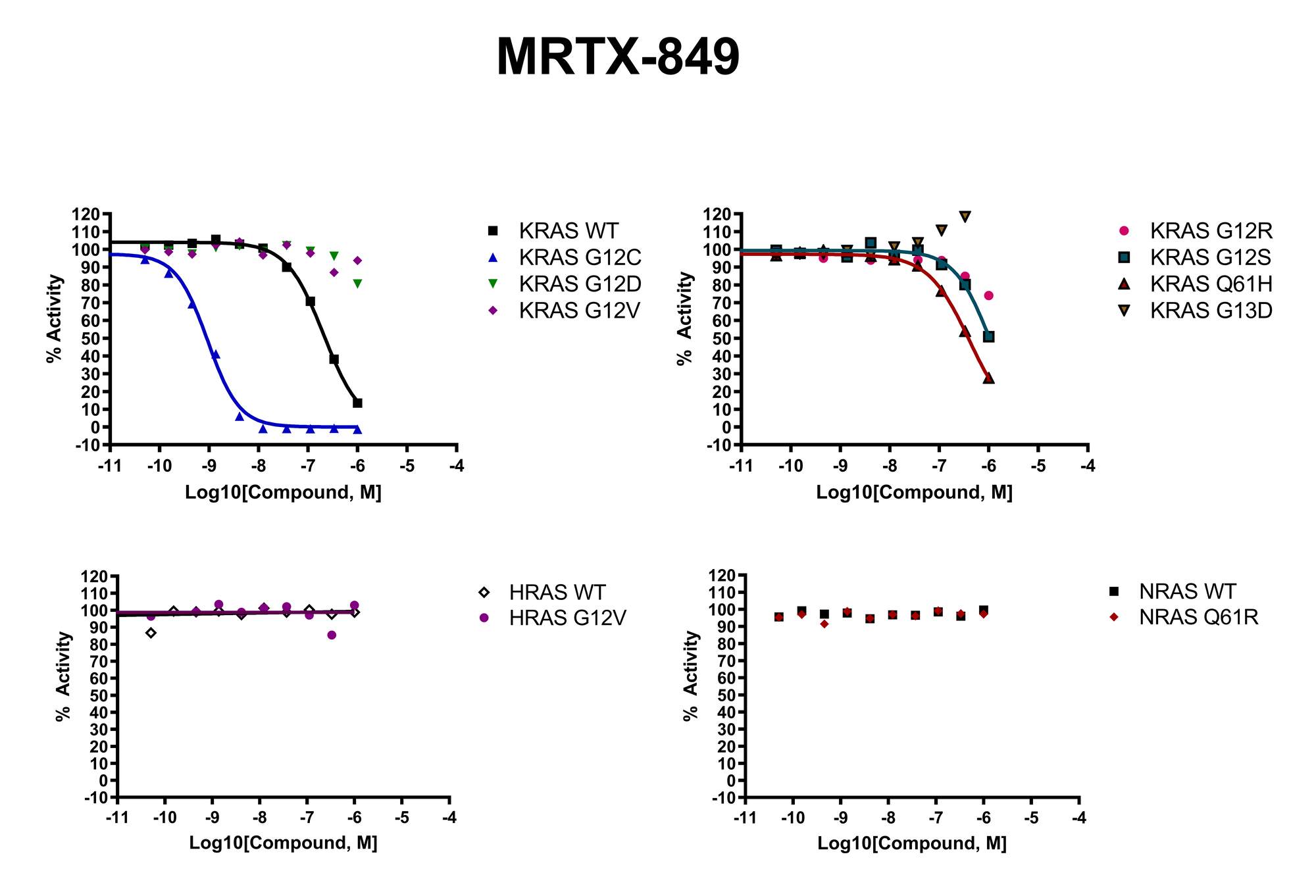
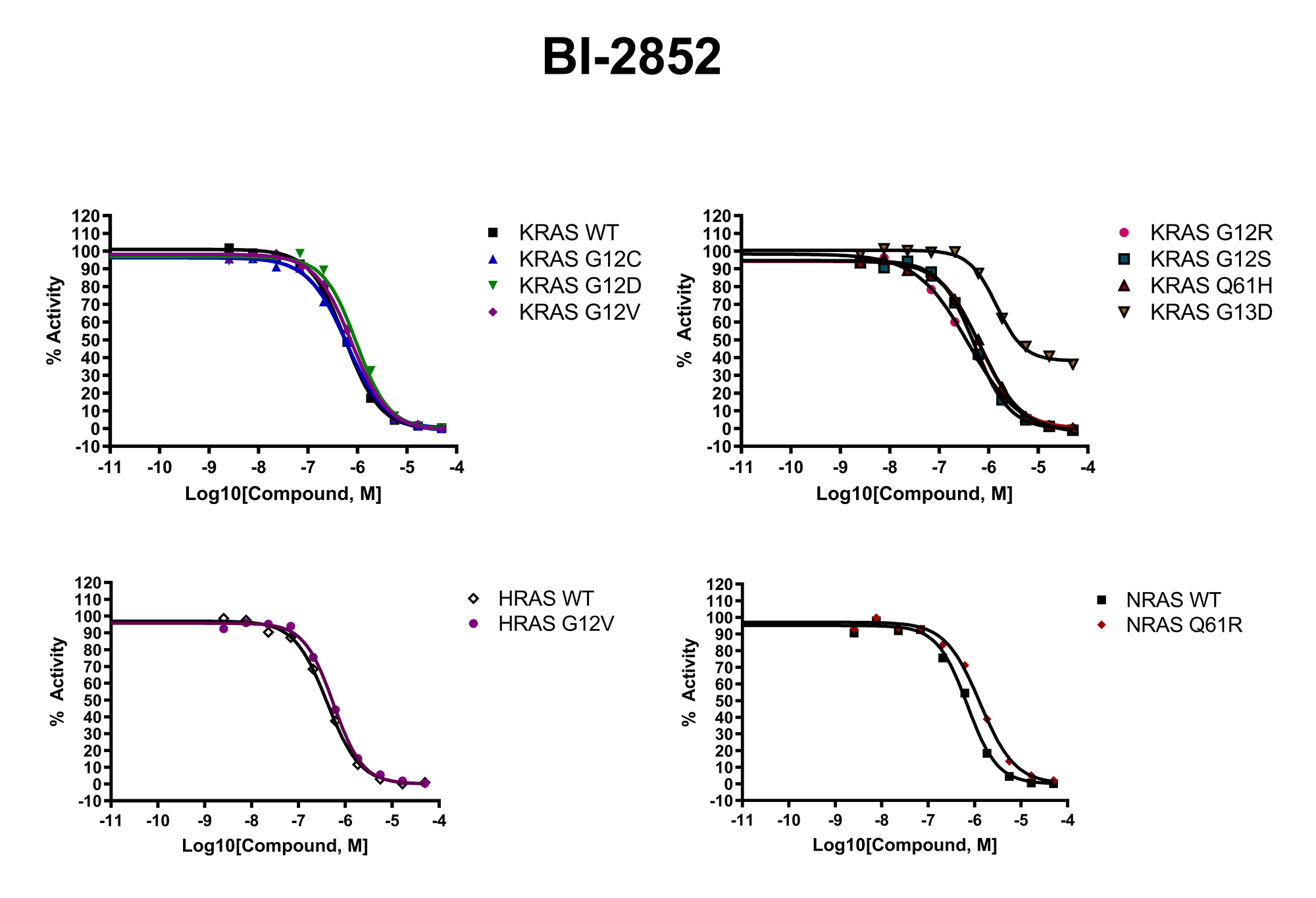
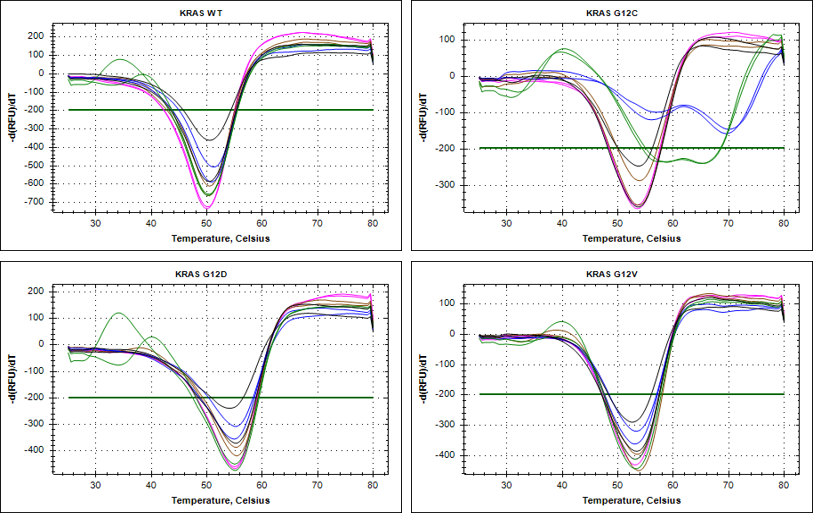
RAS Assays for Drug Discovery
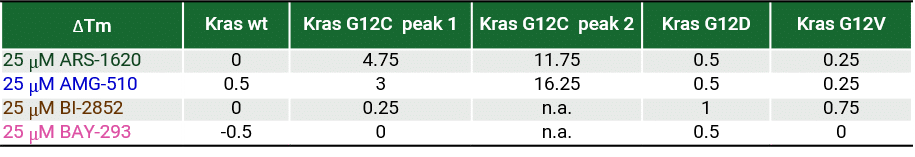
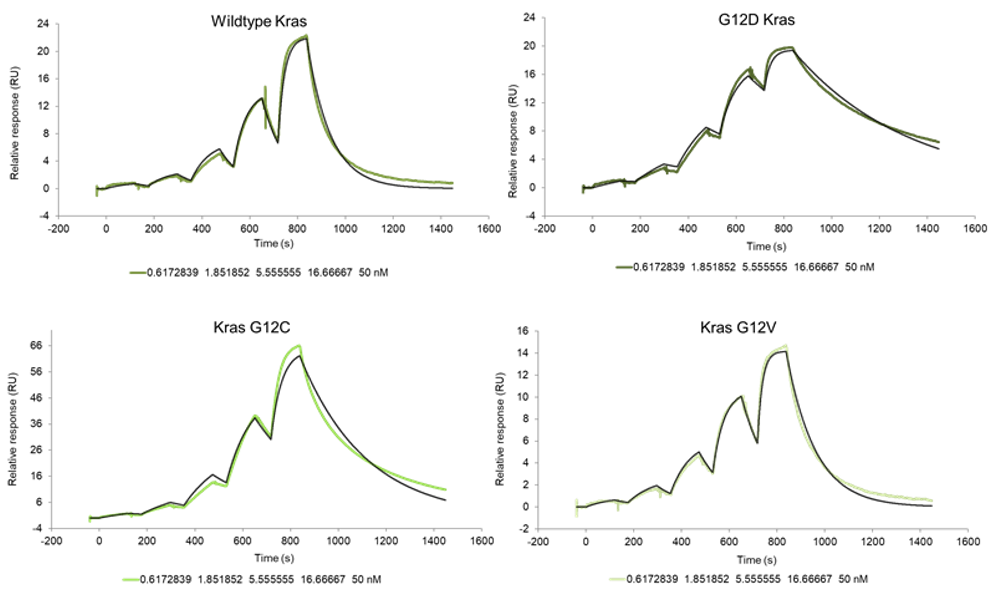
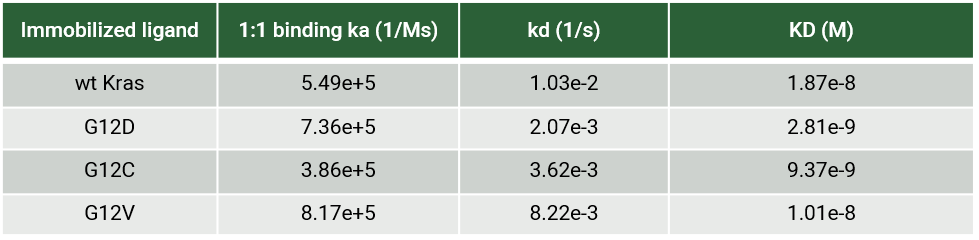
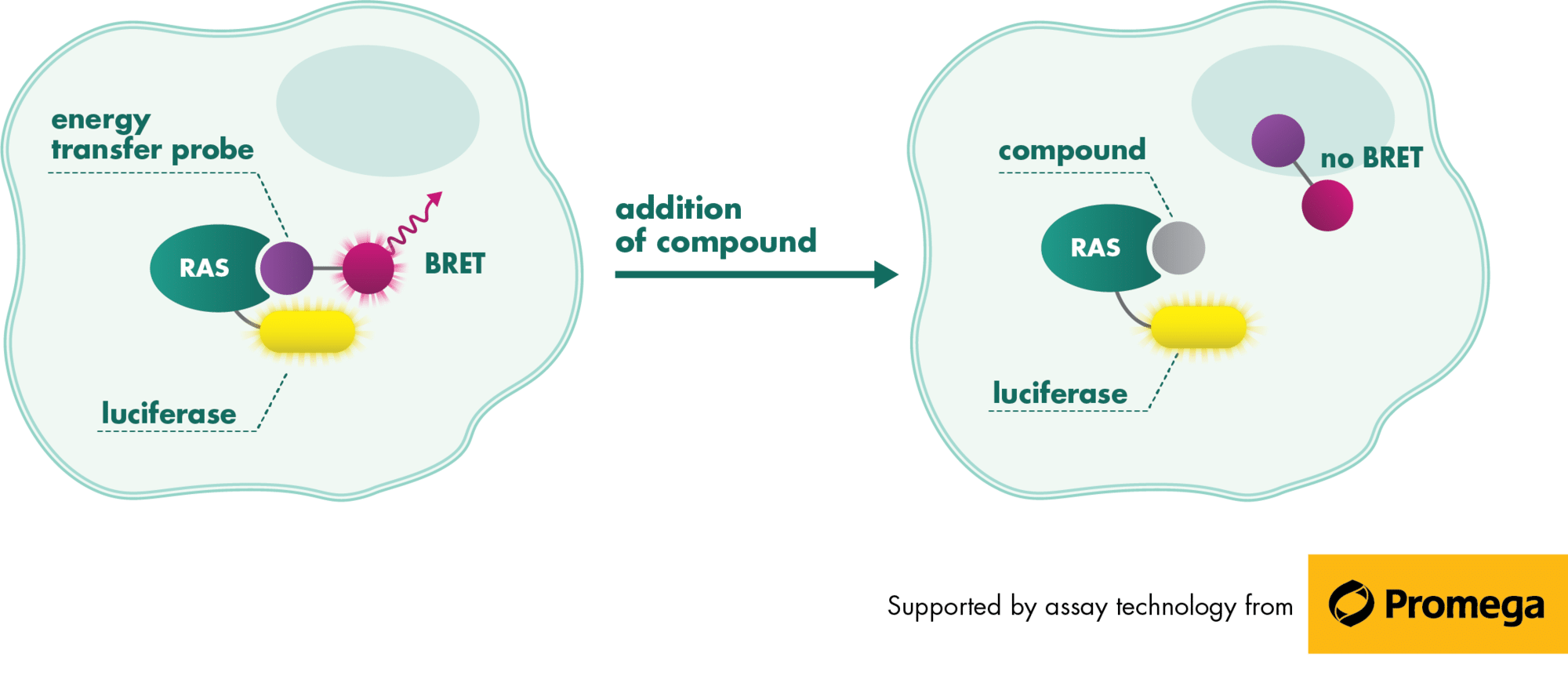
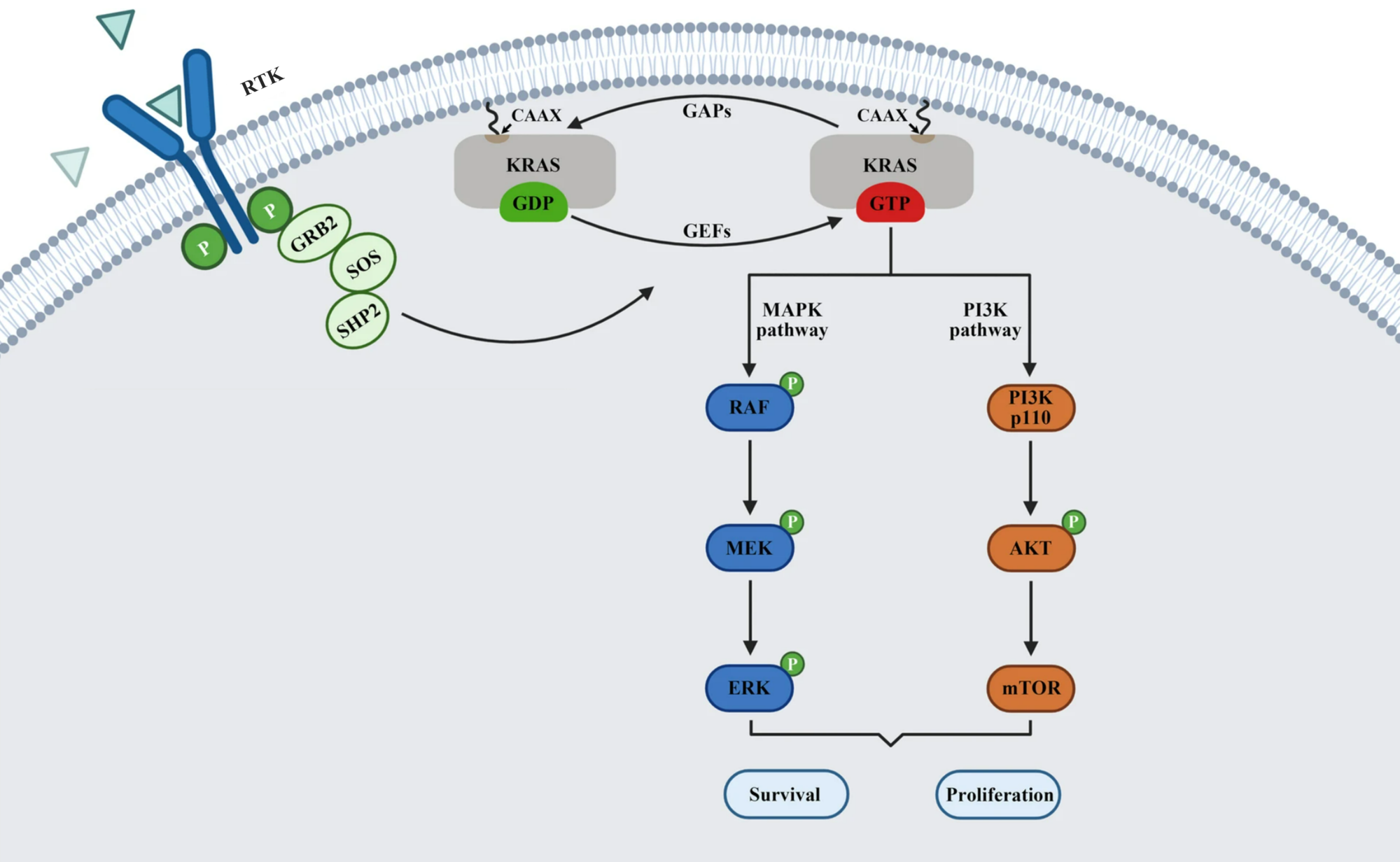
RAS is a small guanosine triphosphate (GTP)-binding protein (GTPase) that is frequently mutated in a large percentage of cancers and is associated with poor disease prognosis. Mutated RAS is locked in the activated GTP bound state and facilitates enhanced RAS signaling in cancer cells. While being a desirable target, the absence of good druggable binding pockets has made modulator compound discovery challenging and unsuccessful.
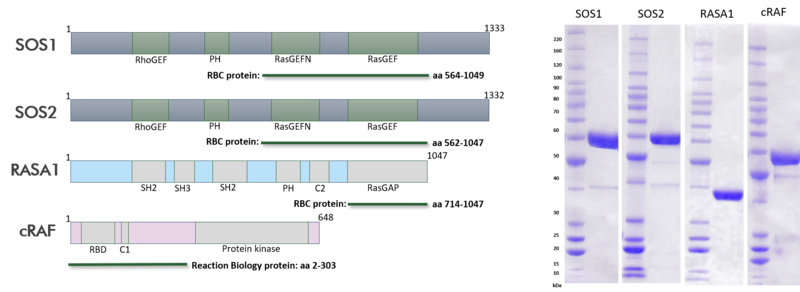
The recent identification of a unique binding pocket and successful inhibition of the KRAS G12C mutant by covalent chemical modifiers have led to the resurgence of interest in the design of inhibitors targeting RAS directly. Alternative efforts are undertaken for the inhibition of interactions of KRAS with exchange factors and effector proteins.
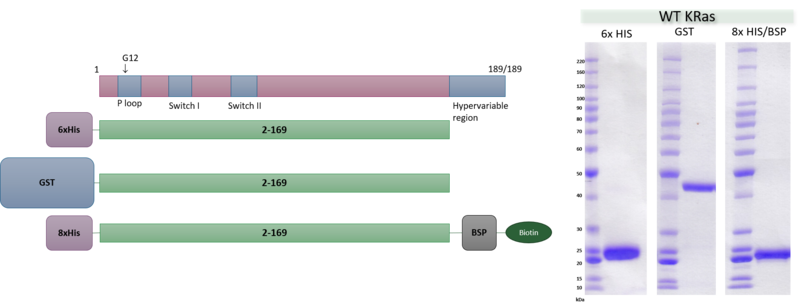
Reaction Biology is committed to quality service in every aspect of our relationships with clients: before, during, and after the study, the assigned business development manager will be just one phone call away to answer any questions. Robust and reproducible assay formats for our RAS assay suite ensure high-quality data.