Histone Deacetylase (HDAC) Assay Services
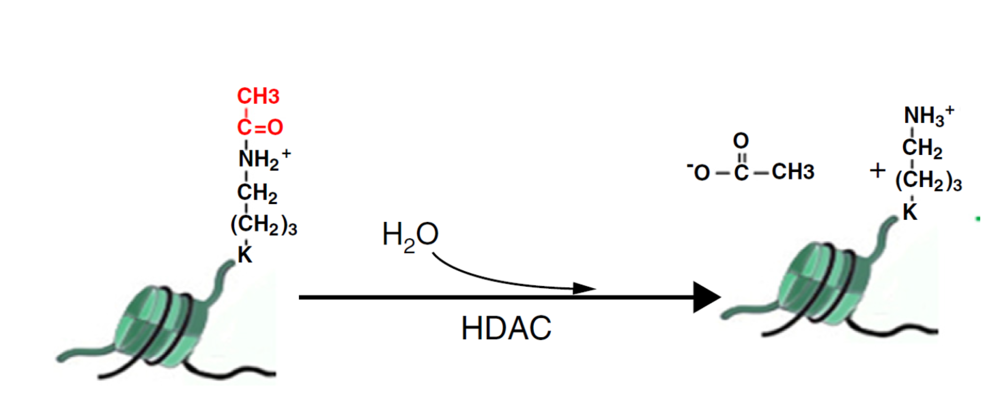
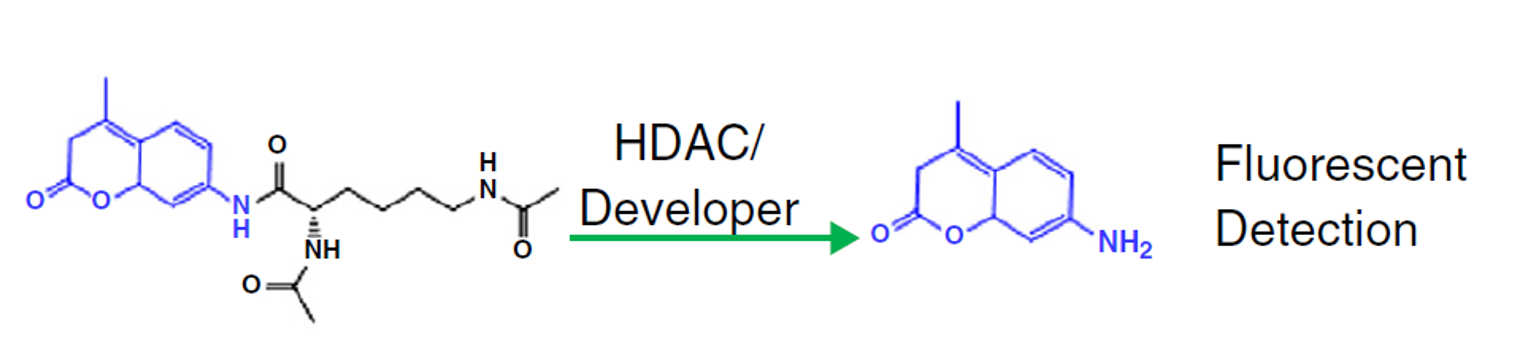
Reaction Biology offers Zn2+-dependent HDAC assays and NAD+-dependent sirtuin assays for compound screening with fluorescence-based activity measurement.
Each HDAC assay and Sirtuin assay is optimized based on its specific substrate.
- Direct measurement of histone deacetylase activity
- Low scale, large scale screening, and high-throughput options
- All HDAC and sirtuin enzymes are produced in-house and available for purchase.
In addition to biochemical assays, we also offer cell-based HDAC assays to measure enzyme activity via the qualification of acetylated histone or tubulin. The assays can be performed via HDAC-Glo, ELISA, or Western blotting.
For drug discovery, the offering comprises all needs of compound screening: primary biochemical HDAC assays, cell-based secondary histone deacetylase assays together with the option of performing HDAC screening via biophysical methods for mechanistic and binding studies.
Histone deacetylase screening is performed at our US facility. A business development manager in your area will be happy to talk to you about your research needs and find out how our team of scientists can support your project best.