Histone Demethylase Assays for Drug Discovery
Histone demethylase assays at Reaction Biology comprise both Jumonji C-domain containing (JmjCs) and lysine-specific demethylase assays (LSD) for compound screening and profiling.
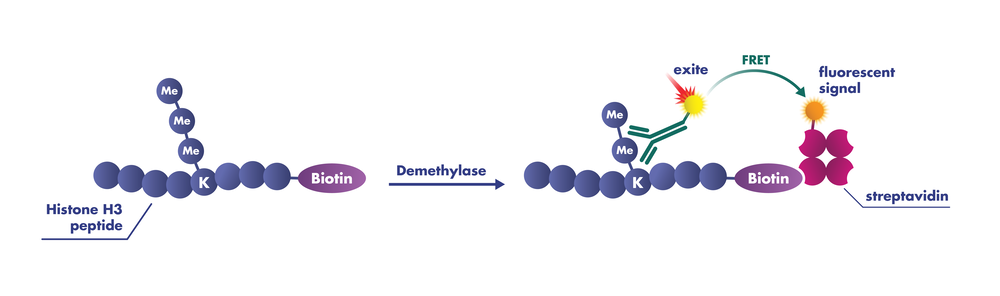
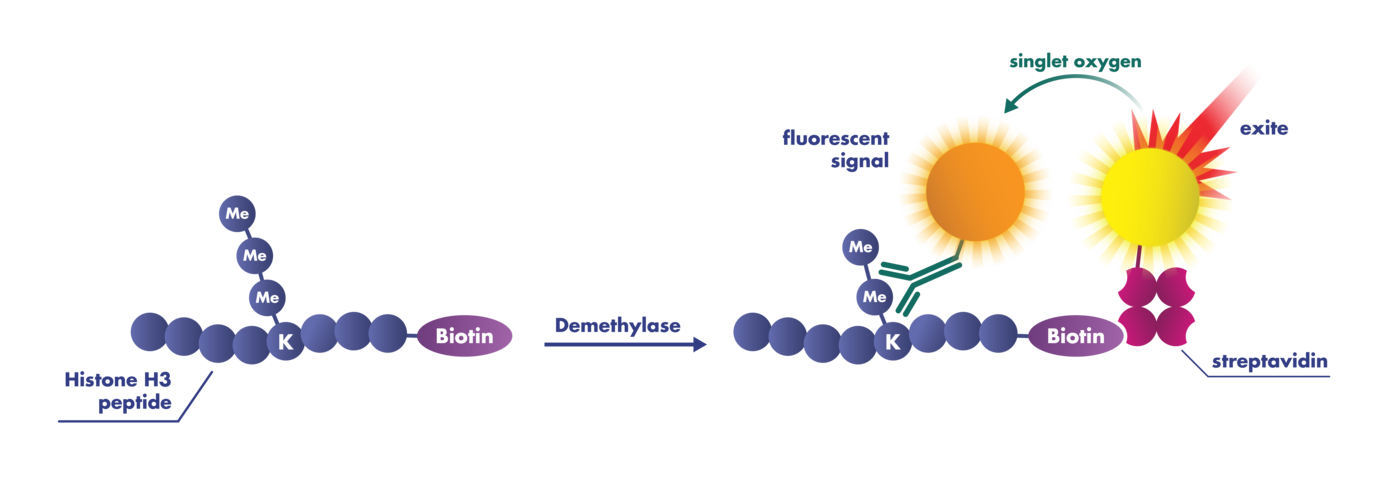
LSDs are flavin-dependent monoamine oxidases that catalyze the demethylation of Kme2 or Kme1, producing peroxide (H2O2) and formaldehyde (H2CO) in the process. JmjCs are Fe(II)/2-oxoglutarate-dependent dioxygenases that use a reactive Fe(IV)-oxoferryl species to catalyze hydroxylation reactions.
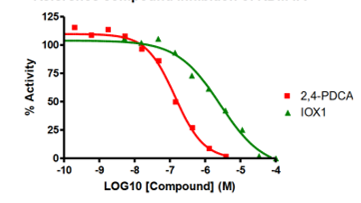
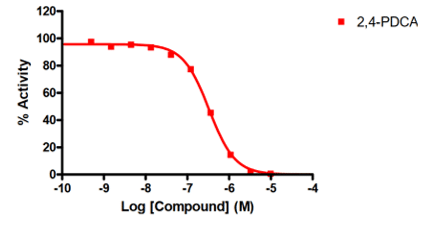
- Demethylase screening is available with three assay formats to measure the potency of inhibitors
- Low scale, large scale, and high-throughput demethylase screening options are available
- All enzymes used in demethylase screening projects are produced in-house and available for purchase.
Reaction Biology performs demethylase assay services at our state-of-the-art facility in Malvern, PA. We have business development managers in multiple locations to easily cater to your assay screening needs. Please ask us for assay development and research collaboration options.
Our histone demethylase assay services have a standard turnaround time of 10 business days. You may talk to the business development manager in your location for expedited assay scheduling and data delivery.