Cell-based Oncology
Phenotypic cellular assays allow the examination of the effects of drug treatment on cells. Reaction Biology offers a large suite of assays including 2D, 3D, or co-culture setups.
Killing basics
Is your drug cytotoxic on tumor cells?
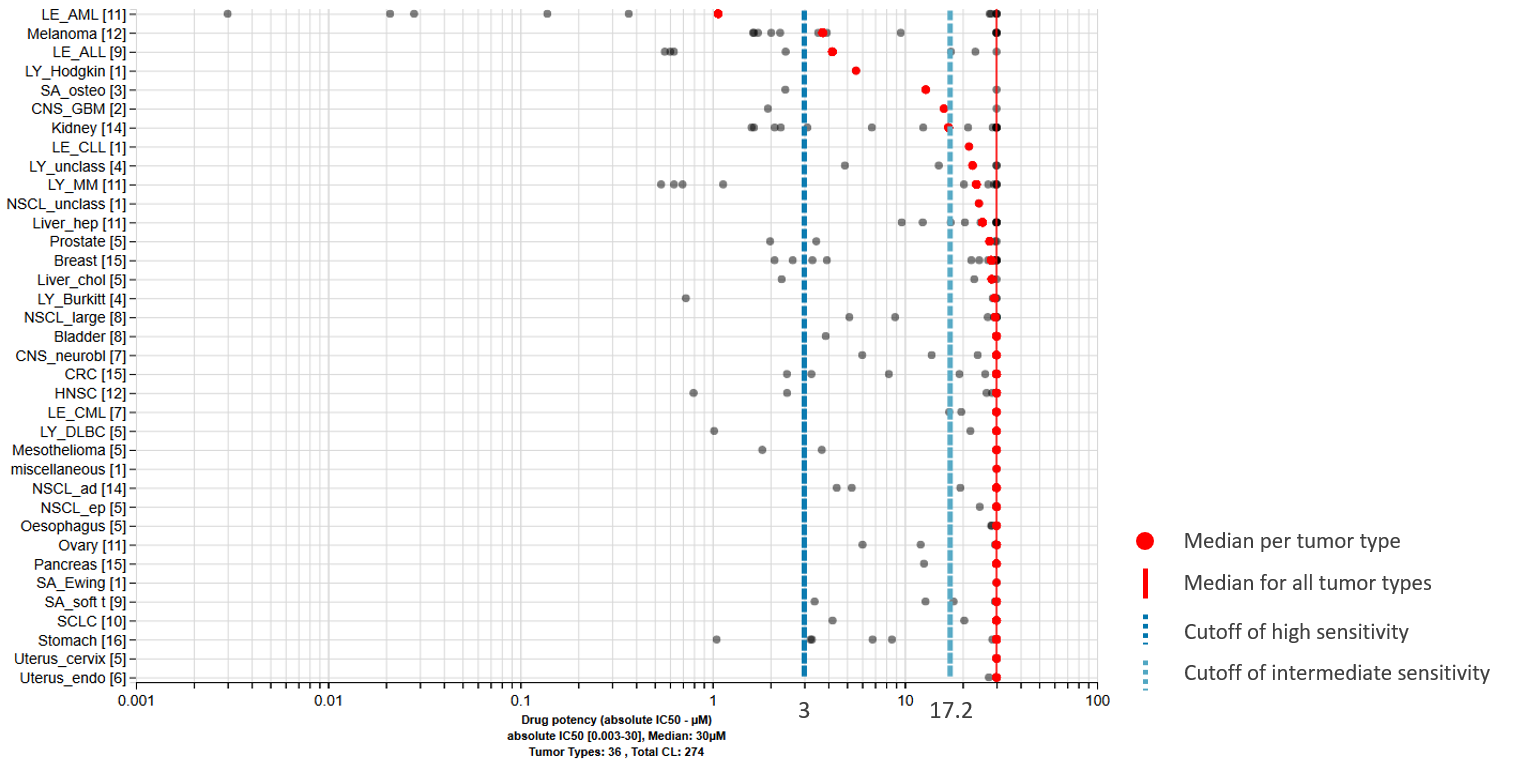
Which tumor cell lines are sensitive to your drug?
Which tumor types are mostly affected by your drug?
Which drug combinations increase the efficacy of your drug?
Killing insight
What is your drug’s kinetics?, or: How fast does your drug kill?
Does your drug induce cell cycle arrest?
Does your drug kill both, tumor and stroma cells?
Tumor cell-specific treatment effects
Is your drug effective on cellular 3D spheroids?
Does your drug inhibit tumor cell migration, a prerequisite of metastasis?
Does your drug stop tumor cells from invading neighboring tissue?
Tumor-specific treatment effects
Does your drug inhibit new blood vessel formation?