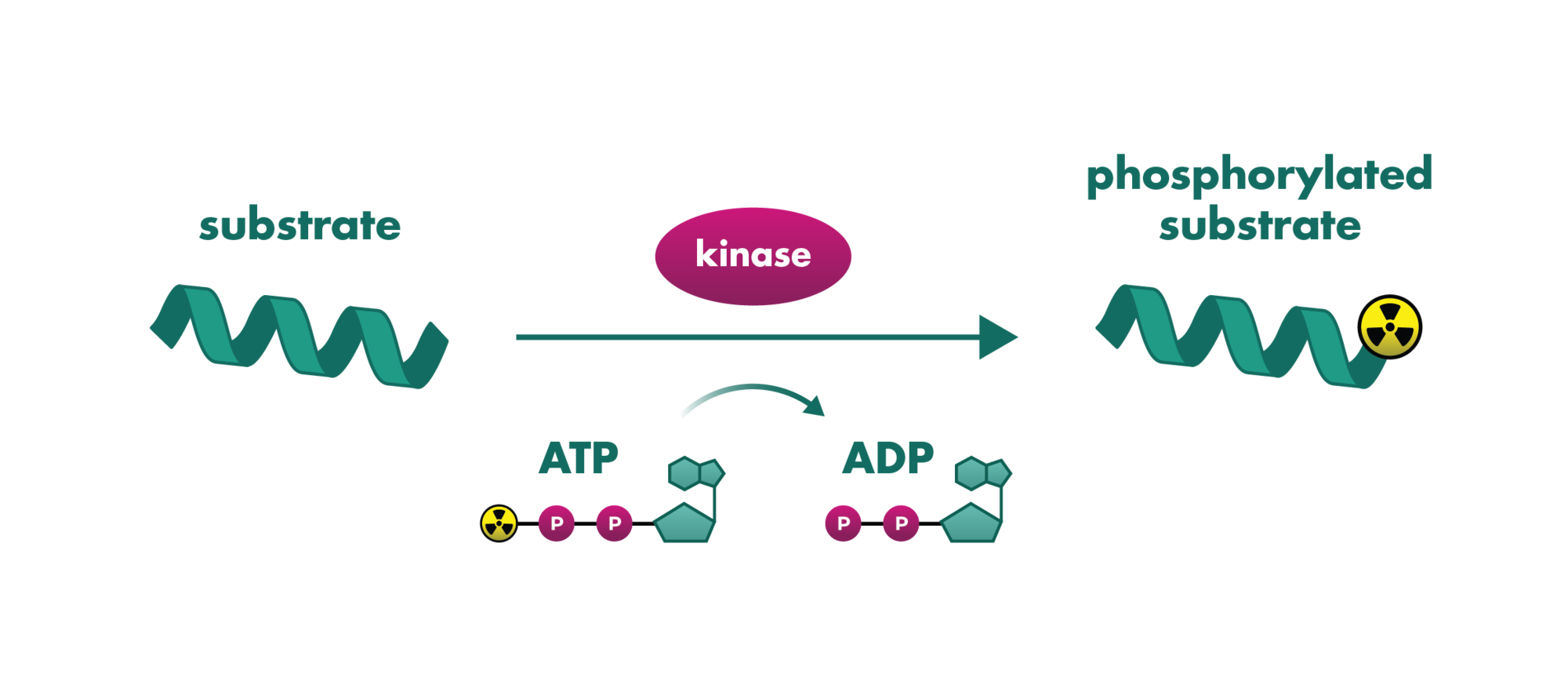
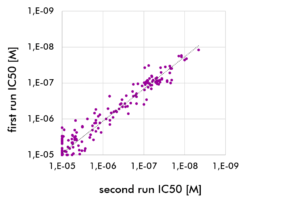
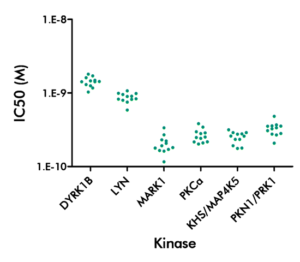
Kinase Panel Screening and Profiling Service
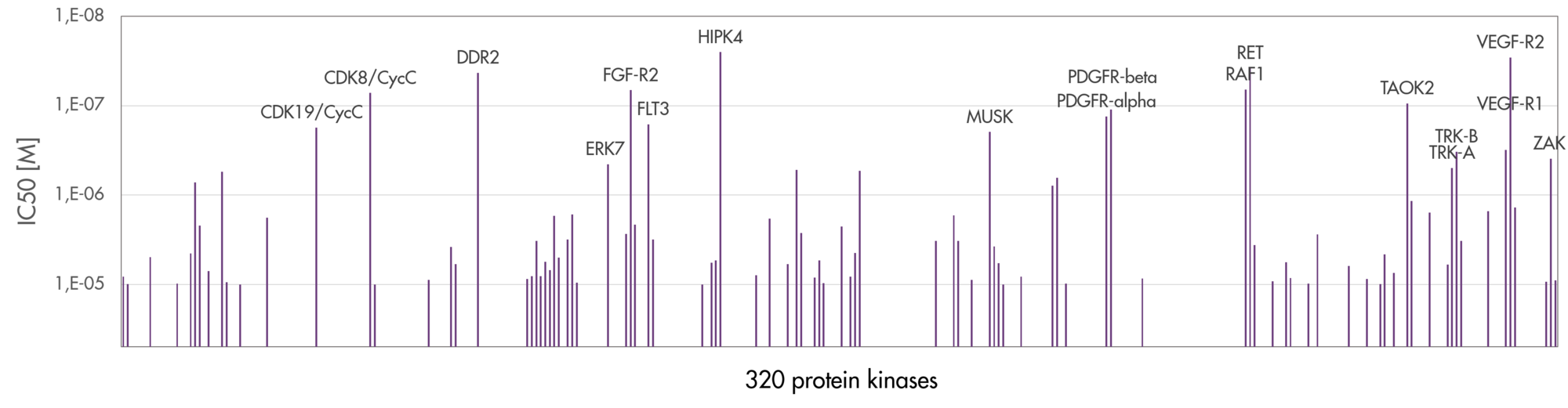
Broad kinase screening is crucial for the identification of selective and potent kinase inhibitors. Determination and improvement of the selectivity of a compound by screening a large part of the kinome are of pivotal importance in the discovery process of novel kinase inhibitors e.g. to reduce the risk of adverse side effects or both, kinase inhibitors and compounds inhibiting non-kinase targets.
- Reaction Biology offers the largest kinase panel for kinase profiling in the industry ensuring the most reliable results for your inhibitor selectivity screening covering most of the human kinome.
- Kinase selectivity profiling is performed on a monthly (³³PanQinase™) or bi-weekly (HotSpot™) schedule, which allows us to offer very competitive pricing.
- HotSpot kinase screening service now available at physiologically relevant 1mM ATP for 340 wild type kinases, in addition to existing standard concentrations of 1μM, 10μM or apparent ATP-Km up to 100μM
- Specialty panels comprise a selection of kinase-specific mutants or our CDK panel.
- Kinase panel reports from the US facility include a free control compound’s IC50 curve.
Reaction Biology’s kinase assays were ranked highest in the Kinase Profiling Trends Survey by HTStec for a reason: Our contract research organization accelerates the drug discovery and development process in two global facilities: one in Pennsylvania, one in Germany. We learn about what our clients want to achieve in their research and match it with our most suitable kinase selectivity profiling service.
As our panels of kinases continue to grow, we consistently demonstrate the most reliable kinase selectivity screening services for clients worldwide based on accurate kinase activity data. Contact us today to discuss how our kinase panel assays can aid your project!